The function of histones goes far beyond packaging DNA into the nucleus. Histones are small proteins that are basic due to their high content of arginine and lysine. They were first identified at the end of the 19th century by the German physiologist Albrecht Kossel1. The combination of two copies of each of the four core histones H2A, H2B, H3, and H4 together with the DNA wound around them form what is known as the nucleosome core particle. Through the assembly of two histone H3/H4 dimers, a stable tetramer is initially formed2,3. Two single histone H2A/H2B dimers4 finally create the histone octamer.
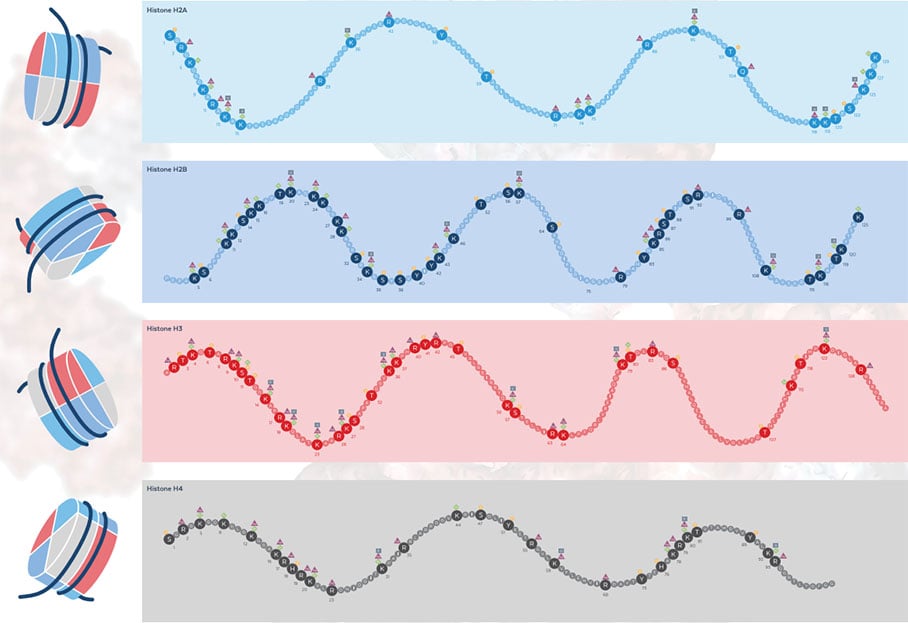
Figure: Comprehensive map of post-translational modifications of histones H2A, H2B, H3, and H4. A full, high-resolution version of this map is available for download.
The first descriptions of post-translationally modified histones were made as early as the mid-1960s. Methylated lysines were detected in the analysis of histones from calf thymus and a short time later, acetylated lysines could also be identified5,6. Since then, other modifications such as arginine methylation, phosphorylation, citrullination, biotinylation, ubiquitination, malonylation, and SUMOylation have been associated with histones (Reviewed in 7). Covalent modifications of specific amino acids of histones form the histone code8, which is the basis for establishing temporally and spatially differentiated chromatin domains (See the figure above for examples).
The best-known modification of terminal histone regions is the methylation of lysine 9 at histone H3 (H3K9). In mammals, for example, it marks facultative or constitutive heterochromatin, i.e. areas of temporal or permanent gene silencing9,10. Histone modifications such as H3K9 methylation are the basis of epigenetics because similar to DNA methylation, the regulation of gene expression is passed on independently of the DNA sequence. Further research into the function of histone modifications will therefore advance our understanding of epigenetics. In addition, altered histone modifications have been implicated in the pathogenesis of an increasing number of diseases, including cancer (Reviewed in 11). Rockland's antibody-based tools are being used by scientists around the world in the pursuit of discovering therapeutic options.
Histone Antibodies By Target
Pan-Specific
| Product | Clonality | Reactivity | Application |
| Lysine Acetylated Antibody | Polyclonal | Broad | WB, IP |
| Lysine Methylated Antibody | Polyclonal | Broad | WB |
| SUMO Antibody | Polyclonal | Broad | WB, ChIP, IP, ELISA |
| Ubiquitin Antibody | Polyclonal | Broad | WB, ELISA |
Histone H3
| Product | Clonality | Reactivity | Application |
| Histone H3 R2me1 Antibody | Polyclonal | Human, C. elegans | WB, Dot Blot |
| Histone H3 R2me2a Antibody | Polyclonal | Human, C. elegans | WB, IF, Dot Blot |
| Histone H3 R2me2s/K4me2 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K4ac Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K4me1 Antibody | Polyclonal | Human | WB, Dot Blot, ELISA |
| Histone H3 K4me2 Antibody | Polyclonal | Human, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K4me3 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 phospho T6 Antibody | Polyclonal | Human, C. elegans | WB, IF, Dot Blot |
| Histone H3 R8me2a Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IHC, IF, ChIP, Dot Blot |
| Histone H3 R8me2s Antibody | Polyclonal | Human, C. elegans | WB, ChIP, Dot Blot |
| Histone H3 K9ac Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K9ac/K14ac Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, Dot Blot |
| Histone H3 K9me1 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K9me2 Antibody | Polyclonal | Human, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K9me3 Antibody | Polyclonal | Human, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 phospho S10 Antibody | Polyclonal | Human, C. elegans | WB, IF, Dot Blot |
| Histone H3 phospho S10/pT11 Antibody | Polyclonal | Human, C. elegans | WB, IF |
| Histone H3 phospho T11 Antibody | Polyclonal | Human | WB, IF, Dot Blot |
| Histone H3 K18ac Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K18me1 Antibody | Polyclonal | Human, Mouse | WB, IF, Dot Blot |
| Histone H3 K18me2 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K18me3 Antibody | Polyclonal | Human, Mouse | WB, IF |
| Histone H3 K23ac Antibody | Polyclonal | Human, Mouse | WB, Dot Blot |
| Histone H3 K23me2 Antibody | Polyclonal | Human, Mouse, Rat, C. elegans | WB, ELISA |
| Histone H3 K27ac Antibody | Polyclonal | Human, Mouse, C. elegans | WB, ChIP, Dot Blot |
| Histone H3 K27me1 Antibody | Polyclonal | Human | WB, IF, ChIP, Dot Blot, ELISA |
| Histone H3 K27me2 Antibody | Polyclonal | Human | WB, IF, ChIP, Dot Blot, ELISA |
| Histone H3 K27me3 Antibody | Polyclonal | Human, Mouse, Rat | WB, IF, Dot Blot |
| Histone H3 phospho S28 Antibody | Polyclonal | Human, Mouse | WB, IF |
| Histone H3 K36ac Antibody | Polyclonal | Human, C. elegans | WB, Dot Blot |
| Histone H3 K36me1 Antibody | Polyclonal | Human, C. elegans | WB, IF, Dot Blot |
| Histone H3 K36me2 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H3 K36me3 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, Dot Blot |
| Histone H3 K37me1 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, Dot Blot |
| Histone H3 K37me2 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, Dot Blot |
| Histone H3 K37me3 Antibody | Polyclonal | Human, C. elegans | WB, IF, Dot Blot |
| Histone H3 K56me1 Antibody | Polyclonal | Human, Mouse | WB, IF, Dot Blot |
| Histone H3 K56me3 Antibody | Polyclonal | Human, C. elegans | WB, IF, Dot Blot |
| Histone H3 K79me1 Antibody | Polyclonal | Human, Mouse, Monkey | WB, IF, ChIP, Dot Blot |
| Histone H3 K79me3 Antibody | Polyclonal | Human, C. elegans | WB, IF, ChIP, Dot Blot |
Histone H4
| Product | Clonality | Reactivity | Application |
| Histone H4 phospho S1 Antibody | Polyclonal | Human | WB, IF, ChIP, Dot Blot |
| Histone H4 R3me1 Antibody | Polyclonal | Human, Mouse | WB, IF |
| Histone H4 K5ac Antibody | Polyclonal | Human, Mouse | WB, IF, ChIP |
| Histone H4 K8ac Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H4 K12ac Antibody | Polyclonal | Human | WB, IF, Dot Blot, Microarray |
| Histone H4 K16ac Antibody | Polyclonal | Human, Mouse | WB, IF, Dot Blot |
| Histone H4 K20me1 Antibody | Polyclonal | Human, Mouse, C. elegans | WB, IF, ChIP, Dot Blot |
| Histone H4 K20me2 Antibody | Polyclonal | Human, C. elegans | WB, IF, ChIP, Dot Blot |
Histone H2A
| Product | Clonality | Reactivity | Application |
| Histone H2A.Zac Antibody | Polyclonal | Broad | WB, IF, ChIP, Dot Blot, ELISA |
| Histone H2AvD phosphoS137 Antibody | Polyclonal | D. melanogaster | WB, IHC, IF, EM, ELISA |
| H2AX phospho S139 Antibody | Polyclonal | Human | WB, ELISA |
References
- Kossel, A., Über einen peptoartigen Bestandteil des Zellkern. Z. Physiol. Chem. 1884, 8, 511–515.
- Kornberg, R. D., & Thomas, J. O. (1974). Chromatin structure; oligomers of the histones. Science (New York, N.Y.), 184(4139), 865–868.
- Roark, D. E., Geoghegan, T. E., & Keller, G. H. (1974). A two-subunit histone complex from calf thymus. Biochemical and biophysical research communications, 59(2), 542–547.
- Kelley R. I. (1973). Isolation of a histone IIb1-IIb2 complex. Biochemical and biophysical research communications, 54(4), 1588–1594.
- ALLFREY, V. G., FAULKNER, R., & MIRSKY, A. E. (1964). ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proceedings of the National Academy of Sciences of the United States of America, 51(5), 786–794.
- MURRAY K. (1964). THE OCCURRENCE OF EPSILON-N-METHYL LYSINE IN HISTONES. Biochemistry, 3, 10–15.
- Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb Perspect Biol. 2015;7(9):a025064.
- Strahl, B. D., & Allis, C. D. (2000). The language of covalent histone modifications. Nature, 403(6765), 41–45.
- Rice, J. C., Briggs, S. D., Ueberheide, B., Barber, C. M., Shabanowitz, J., Hunt, D. F., Shinkai, Y., & Allis, C. D. (2003). Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Molecular cell, 12(6), 1591–1598.
- Wu, R., Terry, A. V., Singh, P. B., & Gilbert, D. M. (2005). Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Molecular biology of the cell, 16(6), 2872–2881.
- Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521.