Immunoprecipitation Technique
Immunoprecipitation (IP) is one of the most widely used approaches for antigen purification and detection. This approach uses antigen-specific antibodies to isolate an antigen of interest from the complex antigen solution and SDS-PAGE to assess the amount and size of an antibody-reactive antigen present in a complex protein mixture. IP enables researchers to measure the molecular weight and the amount of a given protein in the protein mixture, identify the protein activation status and post-translational protein modifications, and capture the protein-binding partners.
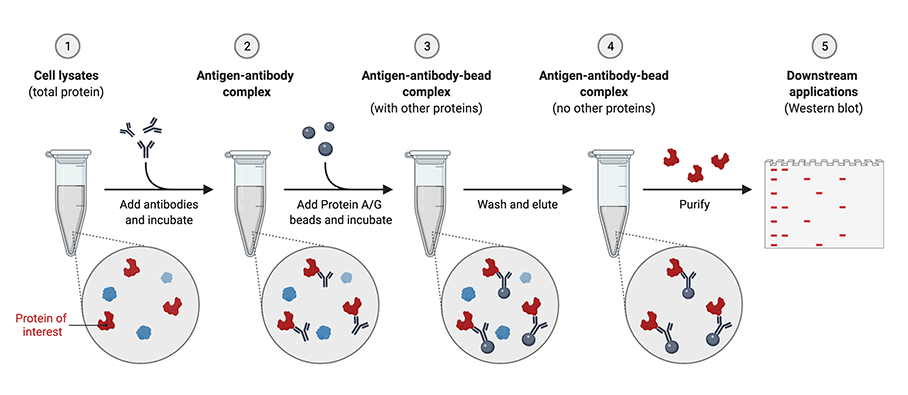
Figure: Immunoprecipitation worklow (Created with BioRender.com)
In general, there are two approaches to capture antigens using antigen-specific antibodies. In the first approach, an antigen-specific antibody (either monoclonal or polyclonal) is pre-immobilized onto a solid support, such as agarose or magnetic beads. Affinity solid matrix is then incubated with a cell lysate that contains the target protein. The immobilized immune complexes are then collected from the lysate, washed, and eluted from the support and analyzed based on the nature of the target antigen. An example of IP with pre-immobilized primary antibodies is IP with anti-FLAG epitope tag applicable to purify protein antigen fused with FLAG eptitope.
In the second IP approach, free, unbound primary antibody is allowed to form immune complexes with the protein of interest presented in the lysate followed by pull-down by using an insoluble affinity support—either protein A, protein G, or protein A/G sepharoses or agaroses conjugated with secondary antibodies such as anti-rabbit IgG agarose conjugates or anti-mouse IgG agarose conjugates. While the pre-immobilized antibody approach is more commonly used for IP, using free antibody to form immune complexes is beneficial if the target protein is present in low concentrations, the antibody has a weak binding affinity for the antigen, or the binding kinetics of the antibody to the antigen are slow.
Traditionally, single proteins have been isolated from cell lysates by IP to investigate the identity, structure, expression, activation, or modification of the individual proteins. IP is also used to study the interaction of a target protein with other proteins or nucleic acids. Variations in the basic IP method provide the flexibility to perform a broad range of applications. For more information of IP please refer to a general method for IP, Chromatin Immunoprecipitation (ChIP), or IP with anti-FLAG antibodies. In general for IP experiment planning, two key components are needed—primary antibodies to form the immune complex with the antigen of interest and a solid support (protein A, protein G, or protein A/G) to pull-down the immune complex.