SDS-PAGE Technique
Polyacrylamide Gel Electrophoresis (PAGE) is one of the most widely used laboratory methods to separate biological macromolecules, such as proteins and nucleic acids. Macromolecules will be differentiated according to their electrophoresis mobility, which is a function of the length, conformation, and charge of the molecule. In general, macromolecules may be run in their native state or in denatured forms. To separate molecules based on their lengths, samples are run in denaturing conditions. For proteins, Sodium Dodecyl Sulfate (SDS) is used to linearize proteins and to negatively charge the proteins. The binding of SDS to the polypeptide chain imparts an even distribution of charge per unit mass. As a result, negatively charged proteins will migrate towards the positive electrode and will be fractionated by approximate size during electrophoresis. This procedure is called SDS-PAGE. Western blotting is the combination of SDS-PAGE and antibody-based detection and is a commonly used antibody application to detect proteins from complex biological mixtures.
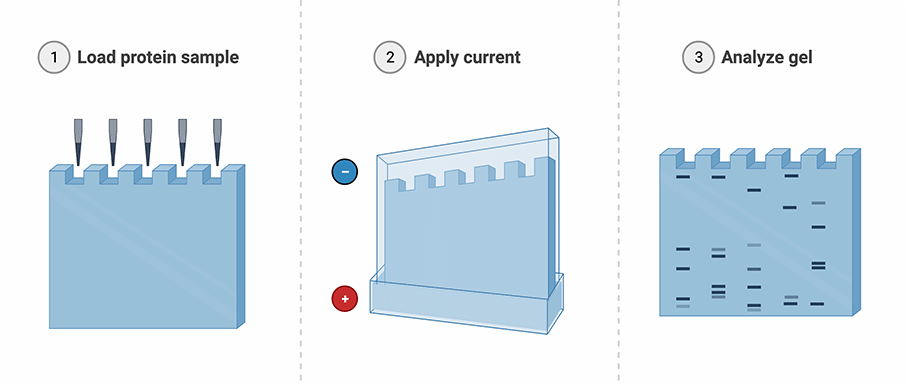
Figure: SDS-PAGE workflow (Created with BioRender.com)
What You'll Need:

Electrophoresis chamber and gel casting kit

Running buffers, Coomassie staining buffers, and destaining buffers

A protein ladder for determining the MW of the protein of interest
- Power supplies: Power supplies convert AC to DC current Electrophoresis chambers: There are various types of chambers sold by suppliers. Be sure to plan ahead and ensure that the electrophoresis chamber that you select fits your SDS-PAGE gel.
- SDS-PAGE gels: You can prepare your own SDS-PAGE gel or purchase them ‘precast’ from commercial sources. Be sure to select a precast gel that fits well into the electrophoresis chamber.
- Proteins samples: Cell lysates or protein mixture can be diluted 1 to 1 using 2X SDS-PAGE Sample Buffer and boiled for 10 minutes. In addition to SDS, a reducing agent such as Dithiothreitol (DTT) or 2-mercaptoethanol is added to reduce disulfide linkages, thus overcoming some forms of tertiary protein folding.
- Protein ladder: Reference Protein Ladder will allow to determine the location of protein of interest based on it molecular size.
- Running buffer: Protein samples loaded on the SDS-PAGE gel will be run in SDS-PAGE Running Buffer.
- Staining buffer: Once the electrophoresis is over, SDS-PAGE gel will be stained in Coomassie Stain Solution. It will take approximately one hour to properly stain the SDS-PAGE gel.
- Destaining buffer: As Coomassie stain also binds to SDS-PAGE gel, a SDS-PAGE Destain Solution is used to destain Coomassie dye from the gel. Protein bands can be visualized by the naked eye or using imaging systems.