pH-Dependent Antibody Engineering
pH-Dependent Antibody Engineering
Reduce on-target, off-tumor side effects in cancer-targeted immunotherapy and CAR-T therapy for solid tumor with the novel, patented approach developed by Abzyme Therapeutics for rapid optimization of pH-dependent antibodies.
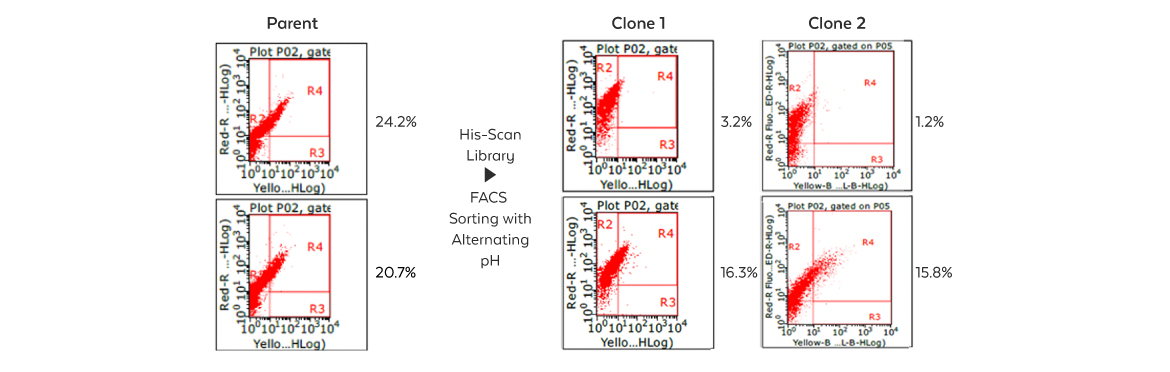
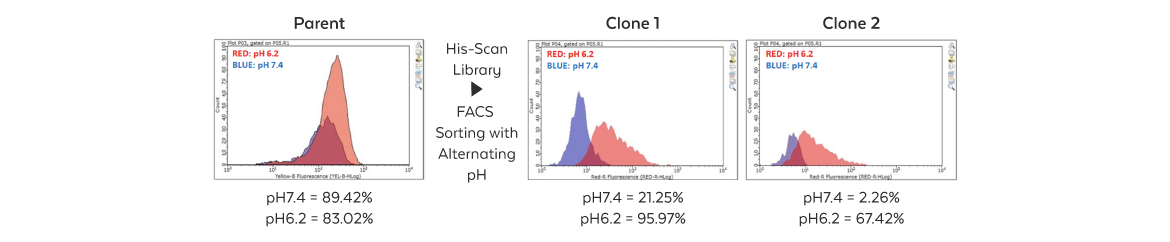
This approach has been successfully applied to isolate antibodies with preferable binding in the acidic tumor microenvironment. It involves designing and synthesizing a His-scan combinatorial library of interacting residues based on the high-resolution structure of the antibody-antigen complex. The library is then cloned into a proprietary surface display vector for rapid screening, allowing low pH binders to be pulled out with a cell sorter.
Benefits of pH-Dependent Antibodies
Monoclonal antibodies are widely used for treatment of various cancers. On-target, off-tumor binding on normal tissues is an obvious toxicity concern. Optimization of specific tumor targeting can be achieved by taking advantage of the extracellular acidity of solid tumors relative to normal tissues. Due to poor vascular perfusion, regional hypoxia, and fermentative glycolysis, the extracellular pH in most solid tumors ranges from 6.0–6.8. In contrast, non-cancerous cells maintain their extracellular pH at physiological levels, 7.3–7.4. Thus, an antibody with high binding affinity at acidic pH, 6.0–6.8, but with reduced or no binding affinity at normal pH will significantly minimize the on-target, off-tumor toxicity.
Tumor-microenvironment—or TME-specific antibodies—with reduced or no on-target, off-tumor binding would find potential application in various forms of antibody- and cell-mediated therapies, including antibody monotherapies, Antibody-Drug Conjugate (ADC) therapy, T cell-engaging immunotherapy, and CAR-T. TME-specific antibodies will reduce on-target, off-tumor adverse effects while allowing dose-escalation.
>
Mediated Toxicity
Safer and more effective biologics for treating cancer by avoiding on-target, off-tumor toxicity
>
Selective Targeting
Quick and efficient ways to select conditionally-active antibodies to improve therapeutic index
>
Clone Isolation
Rapid isolation of desired rare clones from millions of possible variants by flow cytometry
>
Comprehensive Optimization
Real time selection for antibody expressibility, developability, pH-dependence, affinity, specificity, and thermostability
>
Rapid Library Construction
Generate His-scan combinatorial yeast-display library in as little as 3 weeks
>
Multiple Formats
Applicable for single-domain, single-chain Fv, or Fab formats for further multi-specificity engineering
Our Development Process
| Step 1 |
Design The Histidine-scan combinatorial library is designed based on available tertiary structures of the antibody-antigen complex. If the structure is not available, we mutate the residues in the CDR regions in all combinations. |
|
|
3 Weeks
|
Step 2 |
Synthesis His-scan combinatorial library is synthesized. |
| Step 3 |
Construction The surface display library is constructed to express and display the combinatorial antibody library. |
|
| Step 4 |
Isolation Cells that express an antibody binding at acidic pH with reduced or no affinity at a normal pH are isolated by Fluorescence-Activated Cell Sorting (FACS). |
|
| Step 5 |
Sorting Individual clones expressing antibodies are sorted by FACS and then retested for target binding at various pHs and antigen concentrations. |
|
| Step 6 |
Isolation & Sequencing Genes encoding the putative antibody clones are isolated and sequenced. |
|
|
6 Weeks
|
Step 7 |
Expression & Purification pH-dependent antibody candidates are expressed using mammalian expression system. |
| Step 8 |
Characterization Purified pH-dependent antibody candidates are characterized by both immunobiological and cell-based assays |
Case Study
Problem
Bi-specific T-cell engagers (BiTEs) activate T cells through CD3. Using their second binding specificity, BiTEs target the activated T cells to tumor-expressed antigens, thereby bypassing TCR specificity and restriction through Major Histocompatibility Complex (MHC) class I molecules. BiTEs have shown therapeutic efficacy in patients with liquid tumors; however, they do not benefit all patients. To date, no clinical successes of BiTE in solid tumors have been reported.
Solution
A surface His-scanning combinatorial humanized anti-CD3 ScFv library was built based on the existing murine anti-CD3 antibody Okt and screened for pH-dependent antibody clones. Using pH-dependent BiTEs for selective targeting of the tumor microenvironment will address the commonly found BiTE on-target, off-tumor side effect while allowing dose-escalation to improve therapeutic efficacy.
Frequently Asked Questions
-
What antibody format can be developed?
Single domain antibody such as camelid VHH or human VH, single-chain Fv (scFv) and IgG Fab fragments can be expressed and displayed on yeast cell surface. Respectively pH-dependent antibodies can be developed for all these formats.
-
Can you develop pH-dependent for different IgG isotypes?
Yes. pH-dependent antibodies can be first developed in the format of scFv or Fab, then reformatted and expressed in various IgG isotypes.
-
Can you engineer Fc varients that bind to FcRs in a pH-dependent manner?
Yes. Fc can be expressed and displayed on yeast cells surface. Using FcRs as baits one can isolate Fc variants from the combinatorial library that bind to the target FcR in pH-dependent manner.
-
Can you develop antibodies that have high affinity at normal pH 7.4 but onne or reduced activity at acidic pH?
Yes. The same procedure with small modification can allow to select antibody clones that have high affinity at the normal pH and reduced or no binding at lower pH.
-
Can you modulate to improve or reduce binding affinity at certain pH?
Yes. During screening yeast clones expressing antibodies, different antigen concentrations will be used. Based on the antigen concentration, we can isolate high-affinity binder vs low-affinity binder at certain pH.
-
What assays do you use to validate pH-dependent antibodies?
We use various assays such as ELISA, SPR, cytoflow analysis or cell-based functional assays to validate pH-dependent antibodies. Assay will be designed in dependence of antibody functions and your request.
Interested in pH-dependent antibody engineering?
Provide the details of your project by requesting a quote to see how Rockland can assist.